|
Research Discovery and innovation at the DIVA lab is driven by questions at all granlarities of biomedical information including at the genomics, proteomics, phenomics, population, and webscale levels.
|
Discovery |
|||
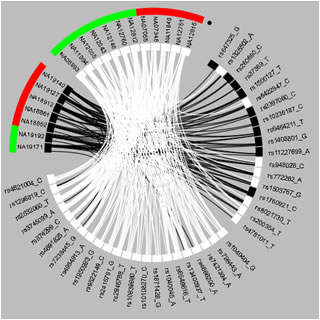 |
Bipartite Visual Analytics of SNP-Subject Data The analysis revealed the role that complementary bipartite visual representations play in helping to generate novel insights into SNP data (Bhavnani et al., 2012). [Distinguished Paper Award] [News]. |
|||
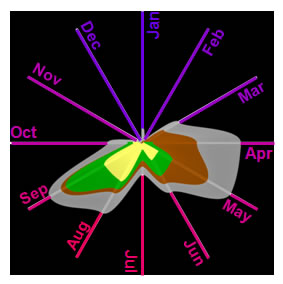 |
Hive Analysis of Asthma Trends |
|||
 |
Circos Ideogram Analysis of Asthma Patient Attributes The analysis revealed complex relationships between asthma molecular and phenotype information, and the complementary role of ideograms to networks (Bhavnani et al., 2011). |
|||
 |
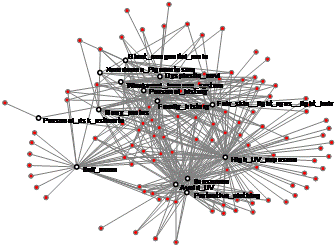
Network Analysis of Asthma Patients and Cytokines The analysis led to insights about a new state-based classification of asthma patients and molecular pathways involved in disease regulation (Bhavnani et al., 2011). [Distinguished Paper Award] |
|||
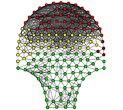 |
Network Analysis of Pregnant Uterine Contractions The analysis explores the use of network modularity to quantify the degree of synchronization of muscle contractions in the uterus, which could help in predicting and preventing premature births (Bhavnani et al., 2011). |
|||
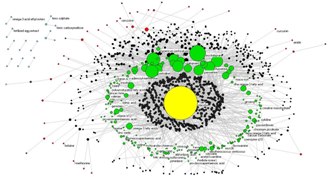 |
Network Analysis of Clinical Trials and Interventions The analysis led to insights about the current state of depression trials, and to a graph-theoretic measure called "Placebo Distance" with applications in comparative effectiveness research (Bhavnani et al., 2010). |
|||
 |
3D Network Analysis of Renal Genes in Immersive CAVE The analysis revealed a new regularity of domain importance that was missed in the 2D network analysis of the same data. The results led to hypotheses for the role played by different CAVE functionalities in enabling new discoveries (Bhavnani et al., 2010). |
|||
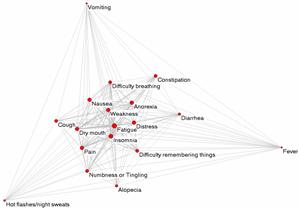 |
Network Analysis of Cancer Patients and Symptoms The results led to the design of algorithms to help clinicians rapidly identify co-occurring symptoms (Bhavnani et al., 2009, Bhavnani et al., 2010). |
|||
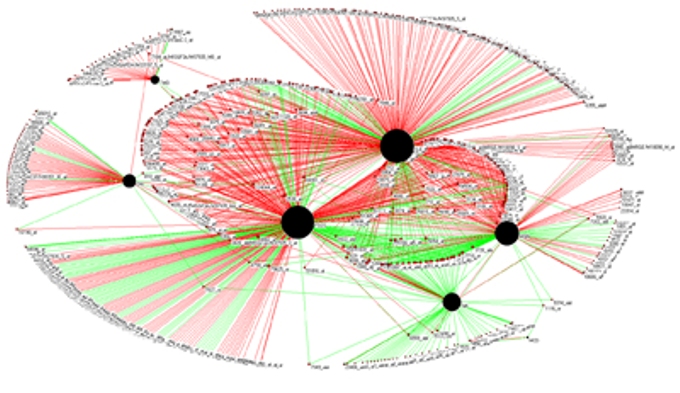 |
Network Analysis of Renal Diseases and Genes |
|||
 |
Network Analysis of Toxic Chemicals and Symptoms The results led to the design of algorithms and interfaces to help first-responders rapidly identify toxic chemicals in emergencies (Bhavnani et al., 2007). [Distinguished Paper Award] |
|||
 |
Scatter of Healthcare Information on the Web The results led to search strategies for finding comprehensive healthcare information, and to the Information Scatter model which proposes how information scatter occurs over time (Bhavnani et al., 2003, Bhavnani, 2005, Adamic et al., 2007, Bhavnani & Wilson, 2010, Bhavnani & Peck, 2010). |
|||
Innovation |
||||
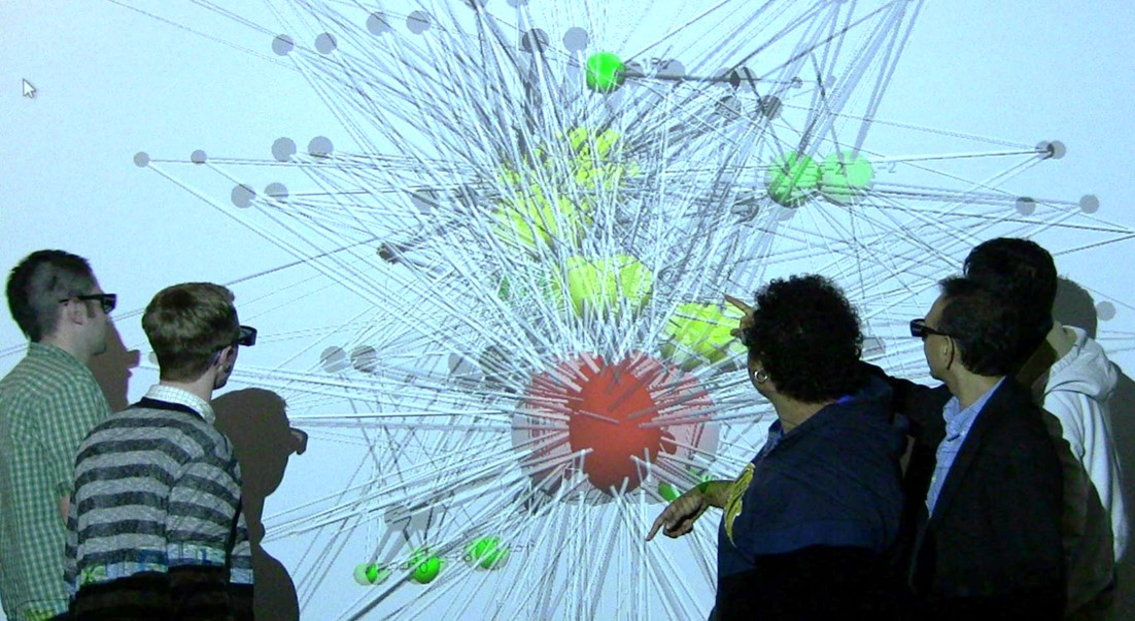 |
Team-Centered Informatics Team-Centered Informatics approaches are designed to help multidisciplinary science teams get an integrated understanding of human biology and disease from large and diverse datasets, with the goal of improving health (Bhavnani et al., 2018). |
|||
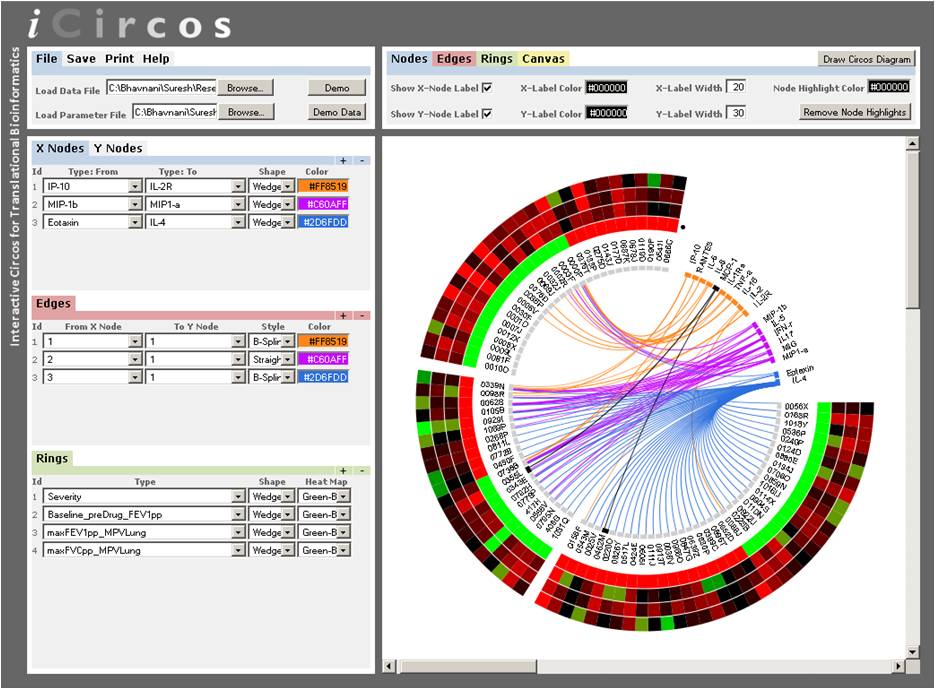 |
iCircos: Visual Analytics for Translational Bioinformatics System to interactively generate Circos representations of bipartite data such as the relationship between patients and cytokines (Bhavnani et al., 2012). |
|||
| Efficient Algorithms for Rapid Diagnosis in Emergencies Extensions of the generalized binary search (GBS) algorithm for identifying groups of objects (e.g., classes of chemicals instead of individual chemicals), and for query noise (e.g., for incorrect inputs due to cognitive stress during emergencies). (Bellala et al., 2010 , Bellala et al., in press ). |
||||
 |
MAIDN: Mining And Interpretation of Diagnostic Networks MAIDN is designed for the rapid identification of toxic chemicals during emergencies. The system significantly reduces the symptoms required to uniquely identify a chemical (Bhavnani et al., 2008, Bhavnani et al., 2008). |
|||
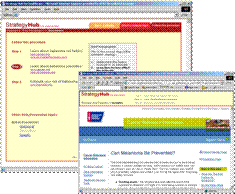 |
Strategy Hub: Web Portal for Finding Comprehensive Cancer Information Users of the Strategy Hub found more comprehensive cancer information in the same amount of time compared to equivalent subjects using MedlinePlus or Google (Bhavnani et al., 2006). |
|||
 |
GoogleBuddy: Social Computing System for Learning and Sharing Search Strategies The results are leading to insights for providing online search knowledge (GROCS Award). |
|||
 |
Strategy-Based Instruction Theory-based, empirically-tested instruction to enable the rapid acquisition of efficient and effective strategies to use complex computer applications. Tested with 400 students in 3 universities including the Nursing School at the University of Michigan (Bhavnani et al., 2001, Bhavnani et al., 2008). |
|||
|
This site maintained by the Institute for Translational Sciences |